Clients with discontiguous regions in their variable-sized chunks may
discover that the library stores their data on disk in a way they don't wish.
The HDF5 library orders the data elements accessed in a dataset according
the row-major ordering that C uses.
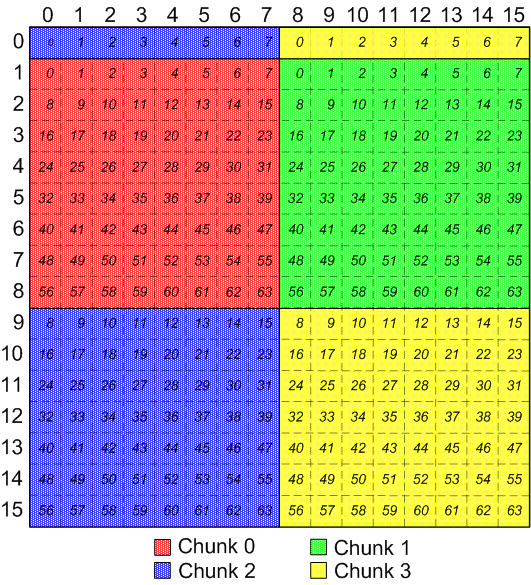
For example, if the chunks for a dataset are discontiguous and
non-overlapping, and are defined like this: (the number inside each element is
the order that the element would be transferred between memory and the file.)
|
Discontiguous, non-overlapping chunks
This is not terribly confusing or suprising for this arrangement of chunks,
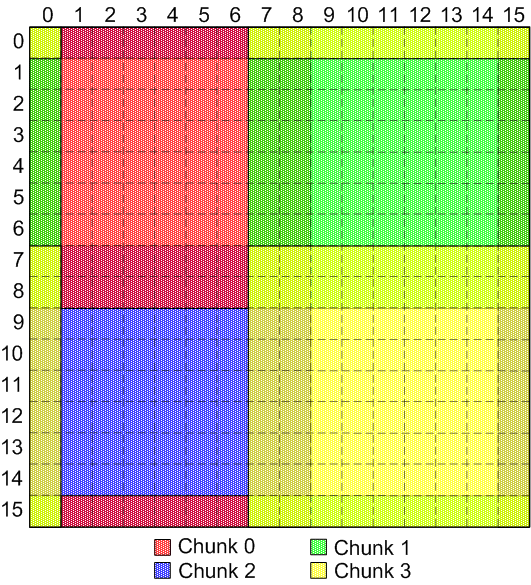
but suppose the client has the following set of overlapping chunks, probably
from a finite-element or finite difference code: (see
here for a view of the individual chunks for this diagram)
|
Discontiguous, overlapping chunks
This corresponds to four 8x8 blocks with single element "ghost zones"
around each block:
| Block # |
"Real" data start location |
"Real" data end location |
| 0 |
(0, 0) |
(7, 7) |
| 1 |
(0, 8) |
(7, 15) |
| 2 |
(8, 0) |
(15, 7) |
| 3 |
(8, 8) |
(15, 15) |
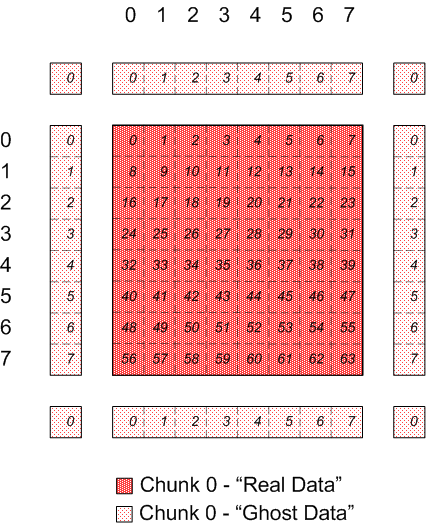
Looking at just chunk 0 and showing the regions with "real" data and "ghost"
data, from the application's point of view:
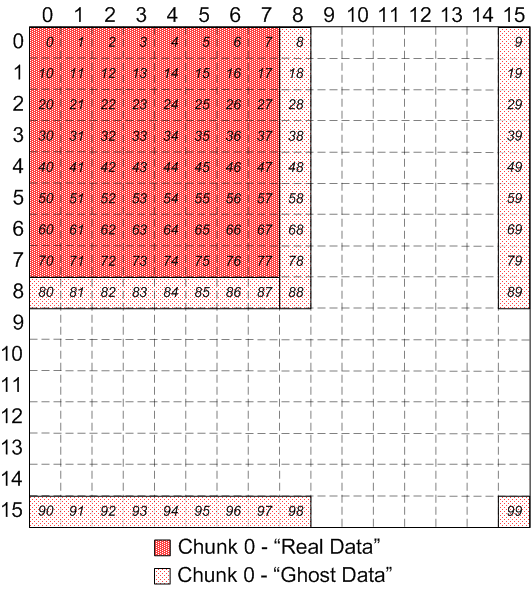
|
Chunk 0 of discontiguous, overlapping chunks
Assume for the purpose of this example, that each process keeps a contiguous
block of "ghost" and "real" data in memory, for their calculations:
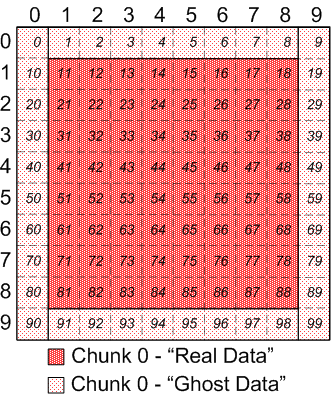
|
Memory view of chunk 0 of discontiguous, overlapping chunks
Following the normal order of traversing the elements in HDF5 will cause the
elements from the ghost and real data to be accessed in an order that the
client probably does not want.
What the client probably wants is this mapping
of elements:
| Memory location |
File location |
| (0, 0) |
(15, 15) |
| (0, 1) |
(15, 0) |
| (0, 2) |
(15, 1) |
| ... |
... |
| (0, 9) |
(15, 8) |
| (1, 0) |
(0, 15) |
| (1, 1) |
(0, 0) |
| (1, 2) |
(0, 1) |
| ... |
... |
What the client is going to get is this mapping:
| Memory location |
File location |
| (0, 0) |
(0, 0) |
| (0, 1) |
(0, 1) |
| (0, 2) |
(0, 2) |
| ... |
... |
| (0, 9) |
(0, 15) |
| (1, 0) |
(1, 0) |
| (1, 1) |
(1, 1) |
| (1, 2) |
(1, 2) |
| ... |
... |
Some potential solutions:
- Don't worry about it - clients don't store ghost zone data around
real data in contiguous blocks in memory:
|
Memory view of chunk 0 of discontiguous, overlapping chunks, stored as separate memory objects
However, this requires the client to make multiple I/O calls to get
the muiltiple blocks in memory out to disk, which is what we are trying
to avoid.
- Ask client to expand dataset's dataspace size by one element in each
direction, so elements on "edges" are actually one element in and there
is room to store ghost zone data without wrapping across dataspace
boundary. This actually ends up mapping back down to identical,
overlapping chunks, but loses the "self description" of the dataset
(i.e. HDF5 thinks the dataset is a 18 x 18 dataspace, not a 16 x 16
dataspace):
(see here for a view of the individual
chunks for this diagram)
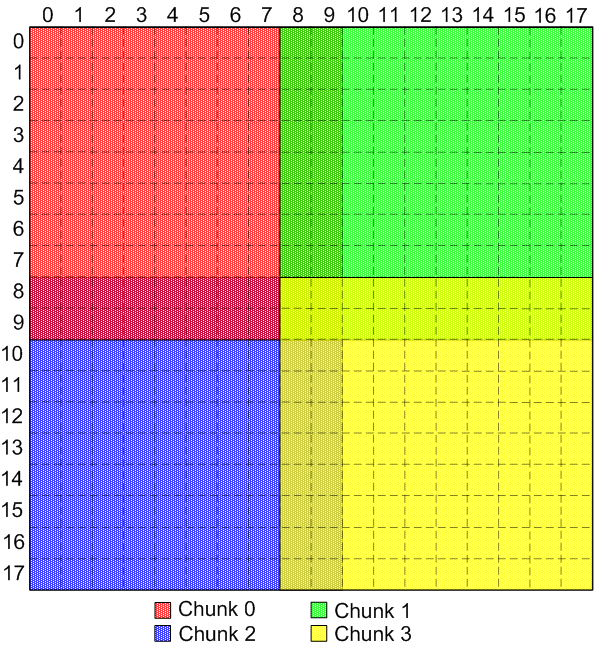
|
Indentical, overlapping chunks
Looking at just chunk 0, you can see that the sequence of elements is
identical to contiguous memory view:
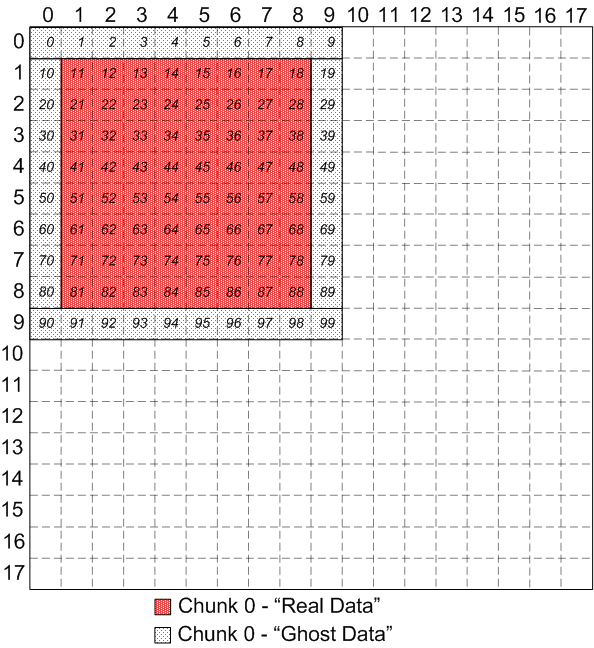
|
Chunk 0 of identical, overlapping chunks
- Allow client to define a "path" through selection and access the
elements in a selection according to the path defined.
Remapping the "path" of elements from chunk 0 of the overlapping,
discontiguous case:
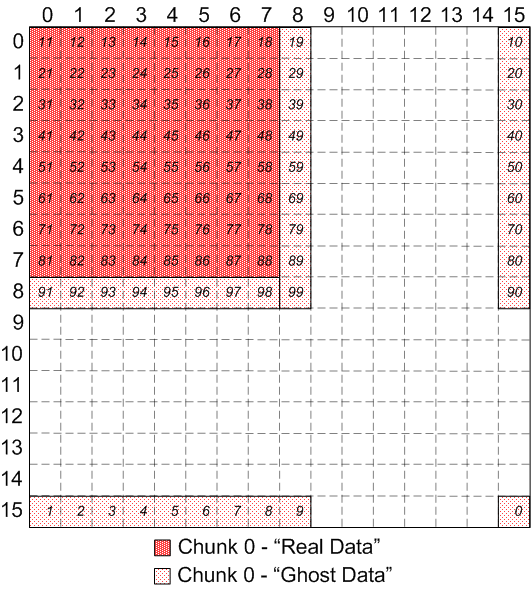
|
Chunk 0 of discontiguous, overlapping chunks, with user-defined path








